|
EESI Tree Generation Tutorial Pipeline Below is an overall picture of the EESI Tree Generation Tutorial Pipeline. Each part of the pipleline is labeled. For information about each part of the label, see the descriptions below. 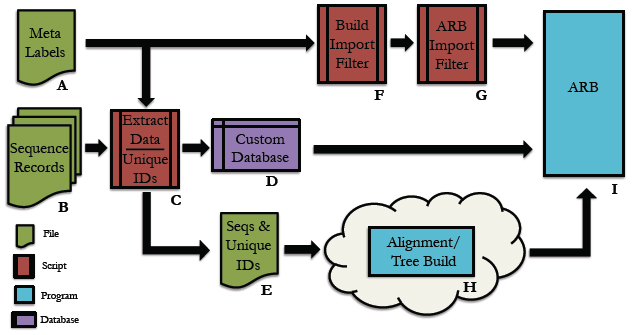 Pipeline Depiction A This is a user supplied text file containing all of the names of the meta-data fields. For each entry in the database, all meta-data will be assigned to its respective name and accessible via this name within the ARB environment. *Note: The order of meta-data names in this file must match the order of the meta-data fields in the custom database. There should be only one meta-data label per line in this text file. See example: metaLabels.txt B Sequence record(s) from which to extract the desired sequence and meta-data information. See example: GenBank File C Python code that extracts information from genbank files. Extensive information can be found in the Data Extraction page. See example: Data Extraction Python File D This is the fasta formatted custom database. Each entry consists of a tab-delimited header followed by a sequence on the next line. To be consistent with the fasta format, the header must start with '>' symbol. A tab is inserted after the carat symbol, which is then followed by the unique id. All meta-data is then filled in, tab-delimited and ordered according to the order of meta-data names provided in the text file described in section A. There must be a tab inserted between the unique id and the start of the first meta-data entry. See example: customDatabase E This is the sequence file for alignment and phylogenetic tree building using any external algorithms and computing resources of choice. It is a fasta-formatted file containing only the unique id in the header. There should be no tabs in the header. See example: sequences.fasta F This is a python script for creating the import filter for your custom database. The metaLabels.txt file described in part A is all that is required by the script to build the filter. The script will output a file called custom_import_filter.ift, which should then be placed in the ARB import filter directory: /arb/lib/import/ See buildFilter.py, custom_import_filter.ift G This is the custom .ift ARB import filter for your database, which is automatically generated by the python script described in part F. The filter must be placed into the ARB import filter directory '/arb/lib/import/'. To import your database you simply follow the typical ARB procedure for creating a new database (shown below). Information on the ARB syntax for the filter may be found here and here. See custom_import_filter.ift 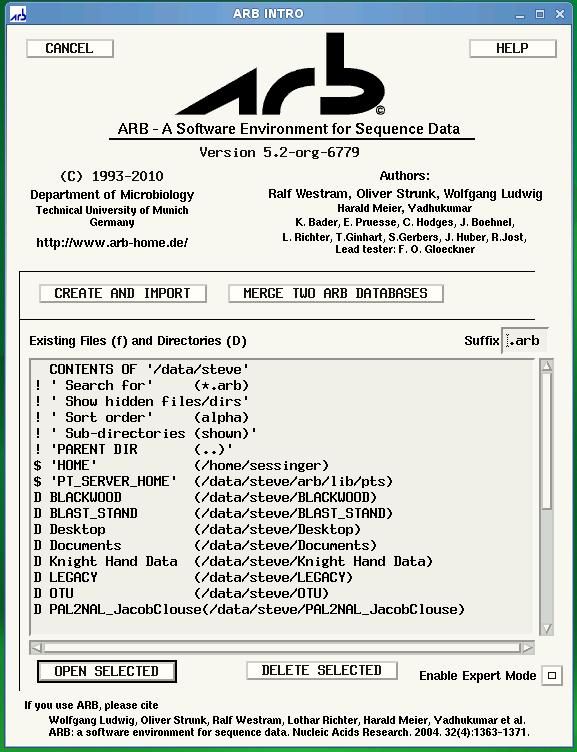 ARB Startup Screen: Choose 'CREATE AND IMPORT' to import your custom database. 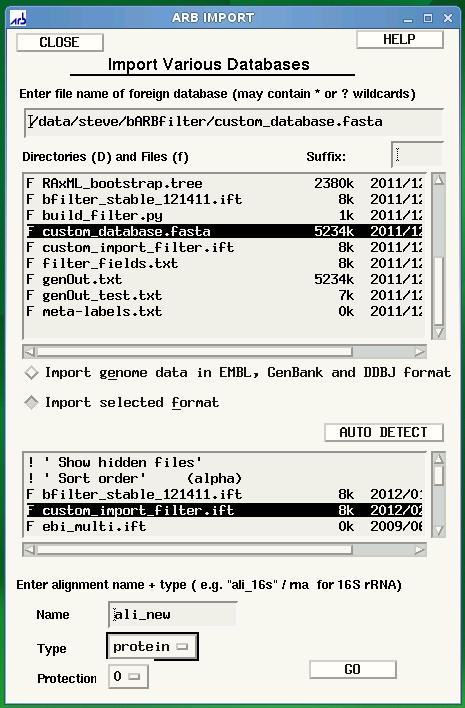 ARB Import screen is displayed after clicking 'CREATE AND IMPORT' shown in previous figure. Select the name of your database under directories and files, then select your custom filter, custom_import_filter.ift. Finally, click GO to begin the import. H The choice of tool for alignment and phylogenetic tree building is at the discretion of the user. For example, you may be interested in building your tree using the maximum likelihood algorithm offered by RAxML, but desire to use external resources due to inadequate internal computational capabilities. The Cipres Science Gateway offers a potential solution for both sequence alignment (e.g. MAFFT) and tree building (e.g. RAxML) using the TeraGrid computing cluster. The user would submit the sequences.fasta file from part E to Cipres and then build a pipeline within Cipres to construct the tree. The tree output from this process would then by imported into ARB using the tree import function found under 'Tree/Tree Admin/Import' within the ARB environment. *Note: The tree file must have the .tree extension in order for ARB to recognize the tree. An example tree file can be found here Cipres Science Portal: http://www.phylo.org/sub_sections/portal 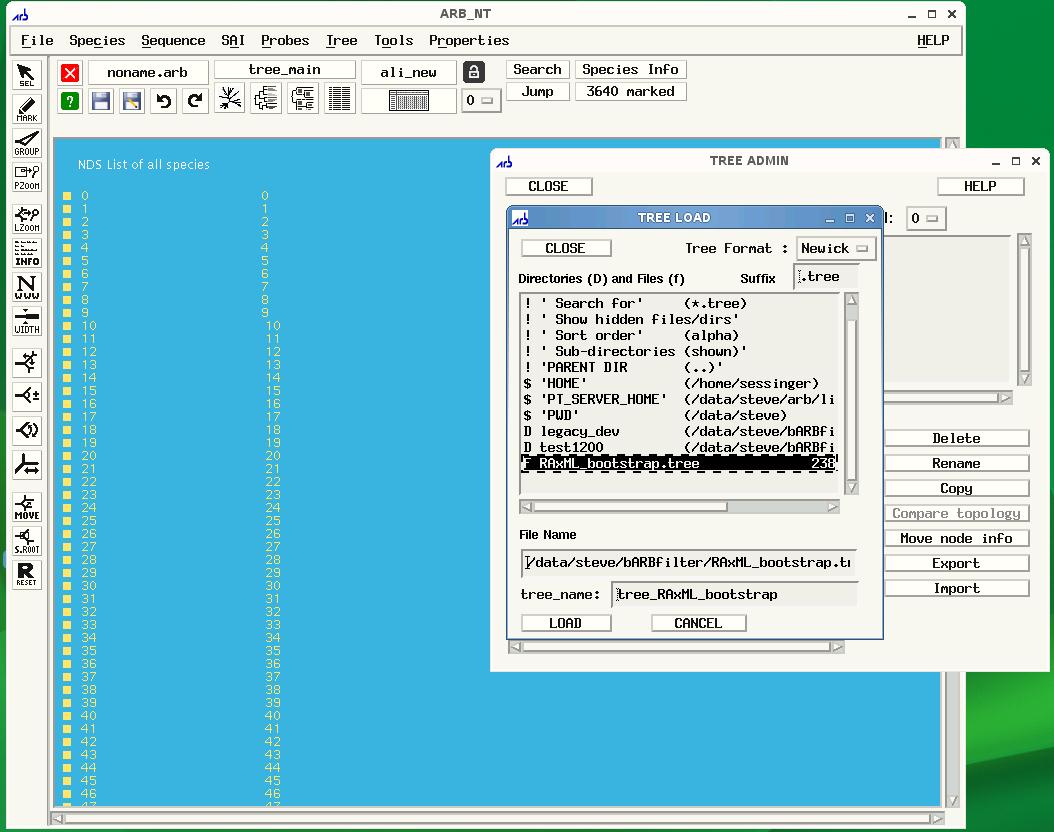 To import your tree, click Tree on the ARB menu, then Tree Admin to open the Tree Admin box and finally, import to open the Tree Load box. Make sure your tree has the .tree extension so that ARB will recognize your file. I This is the ARB program. Information on downloading, installation and use may be found at the ARB website: http://www.arb-home.de |




 © 2006/
site design: e. reichenberger
© 2006/
site design: e. reichenberger